We study gene regulatory mechanisms controlling the timing of animal development using the C. elegans model system. Developmental timing regulators in C.elegans include microRNAs that control the stage-specific expression of key transcription factors. We aim to understand the molecular mechanisms of post-transcriptional gene regulation by microRNAs, and how microRNAs function in regulatory networks affecting development and disease.
Circulating microRNA profiles in human patients with acetaminophen hepatotoxicity or ischemic hepatitis. Ward J, Kanchagar C, Veksler-Lublinsky I, Lee RC, McGill MR, Jaeschke H, Curry SC, Ambros VR. Proc Natl Acad Sci USA. 2014 Aug 19;111(33):12169-74. PMID: 25092309 [PubMed - indexed for MEDLINE] PMCID: PMC4143020.
Abstract:
We have identified, by quantitative real-time PCR, hundreds of miRNAs that are dramatically elevated in the plasma or serum of acetaminophen (APAP) overdose patients. Most of these circulating microRNAs decrease toward normal levels during treatment with N-acetyl cysteine (NAC). We identified a set of 11 miRNAs whose profiles and dynamics in the circulation during NAC treatment can discriminate APAP hepatotoxicity from ischemic hepatitis. The elevation of certain miRNAs can precede the dramatic rise in the standard biomarker, alanine aminotransferase (ALT), and these miRNAs also respond more rapidly than ALT to successful treatment. Our results suggest that miRNAs can serve as sensitive diagnostic and prognostic clinical tools for severe liver injury and could be useful for monitoring drug-induced liver injury during drug discovery.
Legend:
Patient clustering based on earliest miRNA profiles. (A) A clustering of 49 patients (designated by patient ID, are ordered in rows and columns, 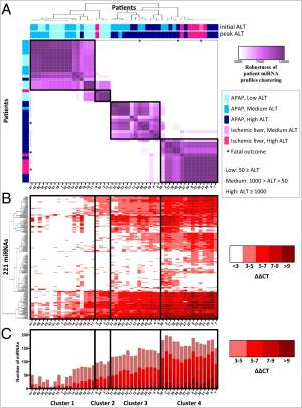 from left to right, top to bottom) was generated based on miRNA profiles from the earliest available sample (within 2 d of hospital admittance). Each patient is designated according to APAP overdose or ischemic hepatitis, initial ALT, peak ALT during hospital stay, and outcome. Each heat map cell corresponds to the number of times that the patient’s miRNA profiles were coclustered out of 10,000 runs of the random k-means algorithm (see details in Materials and Methods). The resulting heat map defines four robust clusters (designated by black boxes, across A, B, and C), denoted as clusters 1–4. (B) The heat map of 221 miRNAs displaying the ΔΔCT = CTSample – Mean CTHealthy. Columns correspond to patients, and rows correspond to unique miRNAs. (C) Plots representing the total number of miRNAs elevated in each patient compared with healthy controls. The colors of the stacked bars correspond to ΔΔCT = CTSample – Mean CTHealthy.
from left to right, top to bottom) was generated based on miRNA profiles from the earliest available sample (within 2 d of hospital admittance). Each patient is designated according to APAP overdose or ischemic hepatitis, initial ALT, peak ALT during hospital stay, and outcome. Each heat map cell corresponds to the number of times that the patient’s miRNA profiles were coclustered out of 10,000 runs of the random k-means algorithm (see details in Materials and Methods). The resulting heat map defines four robust clusters (designated by black boxes, across A, B, and C), denoted as clusters 1–4. (B) The heat map of 221 miRNAs displaying the ΔΔCT = CTSample – Mean CTHealthy. Columns correspond to patients, and rows correspond to unique miRNAs. (C) Plots representing the total number of miRNAs elevated in each patient compared with healthy controls. The colors of the stacked bars correspond to ΔΔCT = CTSample – Mean CTHealthy.
The embryonic mir-35 family of microRNAs promotes multiple aspects of fecundity in Caenorhabditis elegans. McJunkin K, Ambros V. G3 (Bethesda). 2014 Jul 21;4(9):1747-54. doi:10.1534/g3.114.011973. PMID: 25053708[PubMed - indexed for MEDLINE] PMCID:PMC416916.
Abstract:
MicroRNAs guide many aspects of development in all metazoan species. Frequently, microRNAs are expressed during a specific developmental stage to perform a temporally defined function. The C. elegans mir-35-42 microRNAs are expressed abundantly in oocytes and early embryos and are essential for embryonic development. Here, we show that these embryonic microRNAs surprisingly also function to control the number of progeny produced by adult hermaphrodites. Using a temperature-sensitive mir-35-42 family mutant (a deletion of the mir-35-41 cluster), we demonstrate three distinct defects in hermaphrodite fecundity. At permissive temperatures, a mild sperm defect partially reduces hermaphrodite fecundity. At restrictive temperatures, somatic gonad dysfunction combined with a severe sperm defect sharply reduces fecundity. Multiple lines of evidence, including a late embryonic temperature-sensitive period, support a role for mir-35-41 early during development to promote subsequent sperm production in later larval stages. We further show that the predicted mir-35 family target sup-26 (suppressor-26) acts downstream of mir-35-41 in this process, suggesting that sup-26 de-repression in mir-35-41 deletion mutants may contribute to temperature-sensitive loss of fecundity. In addition, these microRNAs play a role in male fertility, promoting proper morphogenesis of male-specific mating structures. Overall, our results demonstrate that robust activity of the mir-35-42 family microRNAs not only is essential for embryonic development across a range of temperatures but also enables the worm to subsequently develop full reproductive capacity.
Legend:
Both somatic gonad and sperm defects contribute to temperature-sensitive loss of fecundity in mir-35-41(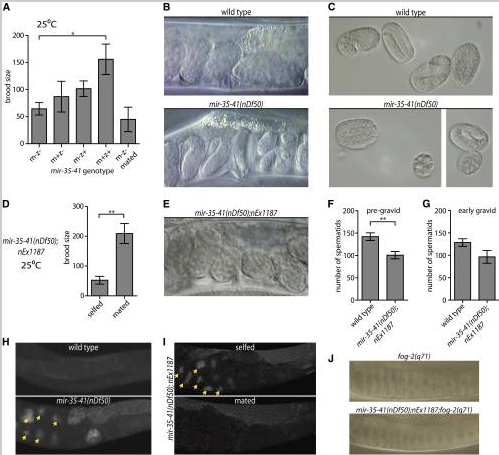 nDf50) hermaphrodites. (A, D, F, G) Mean and SE are plotted. *p-value < 0.05, **p-value < 0.01, two-tailed Student’s t-test. (A) Brood size (y-axis) represents the number of progeny produced per hermaphrodite. m = maternal mir-35-41(nDf50) genotype; z = zygotic genotype. mated = single hermaphrodite was mated to five wild-type males. (B) Proximal uteri of wild-type or mir-35-41(nDf50) hermaphrodites grown at 25°. mir-35-41(nDf50) uteri contain very late-stage embryos, indicative of an egg-laying defective phenotype (Egl). (C) Embryos of wild-type or mir-35-41(nDf50) hermaphrodites grown at 25°. The size and shape of mir-35-41(nDf50) embryos are sometimes abnormal, indicating a defect of somatic gonad architecture. (D) Number of progeny produced by mir-35-41(nDf50) hermaphrodites containing a somatically expressed mir-35 transgene (nEx1187) when selfed or mated to wild-type males. (E) Uteri of mir-35-41(nDf50);nEx1187, indicating that somatic expression of mir-35 rescues the Egl phenotype of mir-35-41(nDf50). (F and G) Number of spermatids per spermatheca in adult hermaphrodites maintained at 25°. Adults were harvested at a (F) pre-gravid or (G) early gravid stage. (H and I) Whole mount DAPI staining in wild-type and mir-35-41(nDf50) hermaphrodites (H) or mir-35-41(nDf50) hermaphrodites containing the mir-35 transgene (nEx1187) when selfed or mated to wild-type males (I). Arrowheads indicate endomitotic oocytes. (J) Proximal gonad of virgin fog-2(q71) females that are otherwise wild-type (top) or containing mir-35-41(nDf50);nEx1187. In both genotypes, oocyte nuclei are pushed close together as oocytes accumulate and stack in the germline.
nDf50) hermaphrodites. (A, D, F, G) Mean and SE are plotted. *p-value < 0.05, **p-value < 0.01, two-tailed Student’s t-test. (A) Brood size (y-axis) represents the number of progeny produced per hermaphrodite. m = maternal mir-35-41(nDf50) genotype; z = zygotic genotype. mated = single hermaphrodite was mated to five wild-type males. (B) Proximal uteri of wild-type or mir-35-41(nDf50) hermaphrodites grown at 25°. mir-35-41(nDf50) uteri contain very late-stage embryos, indicative of an egg-laying defective phenotype (Egl). (C) Embryos of wild-type or mir-35-41(nDf50) hermaphrodites grown at 25°. The size and shape of mir-35-41(nDf50) embryos are sometimes abnormal, indicating a defect of somatic gonad architecture. (D) Number of progeny produced by mir-35-41(nDf50) hermaphrodites containing a somatically expressed mir-35 transgene (nEx1187) when selfed or mated to wild-type males. (E) Uteri of mir-35-41(nDf50);nEx1187, indicating that somatic expression of mir-35 rescues the Egl phenotype of mir-35-41(nDf50). (F and G) Number of spermatids per spermatheca in adult hermaphrodites maintained at 25°. Adults were harvested at a (F) pre-gravid or (G) early gravid stage. (H and I) Whole mount DAPI staining in wild-type and mir-35-41(nDf50) hermaphrodites (H) or mir-35-41(nDf50) hermaphrodites containing the mir-35 transgene (nEx1187) when selfed or mated to wild-type males (I). Arrowheads indicate endomitotic oocytes. (J) Proximal gonad of virgin fog-2(q71) females that are otherwise wild-type (top) or containing mir-35-41(nDf50);nEx1187. In both genotypes, oocyte nuclei are pushed close together as oocytes accumulate and stack in the germline.
An efficient sensitive method for preparing cDNA libraries from scarce biological samples. Sterling, CH, Veksler-Lublinsky, I, Ambros V. Nucleic Acids Res. 2015 Jan;43(1):e1. doi: 10.1093/nar/gku637. Epub 2014 Jul 23. PMID:25056322 [PubMed - indexed for MEDLINE] PMCID:PMC4288208
Abstract:
The preparation and high-throughput sequencing of cDNA libraries from samples of small RNA is a powerful tool to quantify known small RNAs (such as microRNAs) and to discover novel RNA species. Interest in identifying the small RNA repertoire present in tissues and in biofluids has grown substantially with the findings that small RNAs can serve as indicators of biological conditions and disease states. Here we describe a novel and straightforward method to clone cDNA libraries from small quantities of input RNA. This method permits the generation of cDNA libraries from sub-picogram quantities of RNA robustly, efficiently and reproducibly. We demonstrate that the method provides a significant improvement in sensitivity compared to previous cloning methods while maintaining reproducible identification of diverse small RNA species. This method should have widespread applications in a variety of contexts, including biomarker discovery from scarce samples of human tissue or body fluids.
Legend:
Overview of 3-day generation of cDNA libraries. (A) On the first day, total RNA is ligated to a 3′ adapter and cDNA is generated by reverse transcription by tandem reactions in a single tube, RNA is degraded and cDNAs are isolated by ethanol precipitation. (B)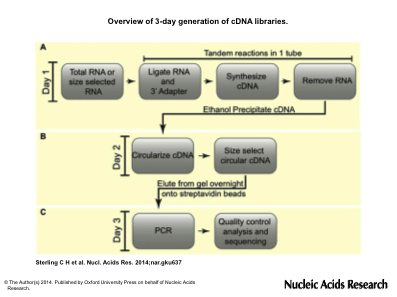 On the second day, cDNAs are circularized, size selected by gel fractionation and eluted overnight in the presence of streptavidin beads. (C) PCR is done on bead-bound purified cDNAs to generate templates ready for high-throughput sequencing.
On the second day, cDNAs are circularized, size selected by gel fractionation and eluted overnight in the presence of streptavidin beads. (C) PCR is done on bead-bound purified cDNAs to generate templates ready for high-throughput sequencing.
Abstract:
microRNAs function in diverse developmental and physiological processes by regulating target gene expression at the post-transcriptional level. ALG-1 is one of two Caenorhabditis elegans Argonautes (ALG-1 and ALG-2) that together are essential for microRNA biogenesis and function. Here, we report the identification of novel antimorphic (anti) alleles of ALG-1 as suppressors of lin-28(lf) precocious developmental phenotypes. The alg-1(anti) mutations broadly impair the function of many microRNAs and cause dosage-dependent phenotypes that are more severe than the complete loss of ALG-1. ALG-1(anti) mutant proteins are competent for promoting Dicer cleavage of microRNA precursors and for associating with and stabilizing microRNAs. However, our results suggest that ALG-1(anti) proteins may sequester microRNAs in immature and functionally deficient microRNA Induced Silencing Complexes (miRISCs), and hence compete with ALG-2 for access to functional microRNAs. Immunoprecipitation experiments show that ALG-1(anti) proteins display an increased association with Dicer and a decreased association with AIN-1/GW182. These findings suggest that alg-1(anti) mutations impair the ability of ALG-1 miRISC to execute a transition from Dicer-associated microRNA processing to AIN-1/GW182 associated effector function, and indicate an active role for ALG/Argonaute in mediating this transition.
Legend:
alg-1(anti) mutations affect functions of many microRNAs and exhibit phenotypes more severe than those of alg-1(0).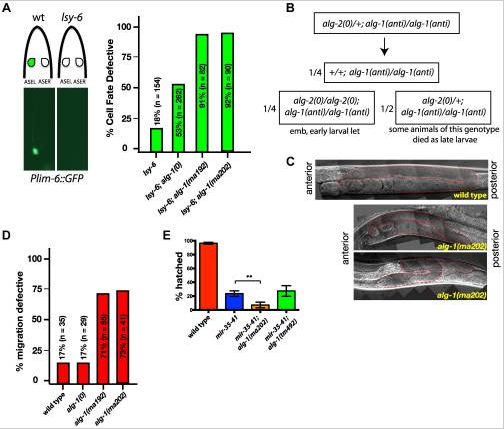 (A) Plim-6::gfp expression marks ASEL neuronal cell fate in wild type and mutant animals. lsy-6(ot150) mutants lack the Plim-6::gfp expression in the ASEL neurons some of the time. This phenotype is enhanced by the loss of alg-1 and even more so in the presence of either of the two alg-1(anti) mutations. All strains carry lin-31 mutation in order to suppress alg-1(anti) vulval bursting phenotypes by non-heterochronic methods. (B) Combination of alg-1(anti) and alg-2(0) mutations results in embryonic lethality (emb) and early larval lethality (let), with some late larval lethality present in alg-2(0)/+; alg-1(anti)/alg-1(anti) mutant animals. (C, D) Distal tip cell migration phenotypes of wild type and mutant animals. alg-1(anti) mutant animals display defects affecting all three phases of gonad migration. In (C) gonads are marked by a dashed line. (E) alg-1(anti), but not alg-1(0) mutation enhances the embryonic lethality of mir-35–41 mutants. (**p<0.001). All strains carry lin-31(lf) and col-19::gfp in the background. The lin-31 mutation is present in all strains in order to suppress alg-1(anti) vulval bursting phenotypes by non-heterochronic methods. n=number of animals scored.
(A) Plim-6::gfp expression marks ASEL neuronal cell fate in wild type and mutant animals. lsy-6(ot150) mutants lack the Plim-6::gfp expression in the ASEL neurons some of the time. This phenotype is enhanced by the loss of alg-1 and even more so in the presence of either of the two alg-1(anti) mutations. All strains carry lin-31 mutation in order to suppress alg-1(anti) vulval bursting phenotypes by non-heterochronic methods. (B) Combination of alg-1(anti) and alg-2(0) mutations results in embryonic lethality (emb) and early larval lethality (let), with some late larval lethality present in alg-2(0)/+; alg-1(anti)/alg-1(anti) mutant animals. (C, D) Distal tip cell migration phenotypes of wild type and mutant animals. alg-1(anti) mutant animals display defects affecting all three phases of gonad migration. In (C) gonads are marked by a dashed line. (E) alg-1(anti), but not alg-1(0) mutation enhances the embryonic lethality of mir-35–41 mutants. (**p<0.001). All strains carry lin-31(lf) and col-19::gfp in the background. The lin-31 mutation is present in all strains in order to suppress alg-1(anti) vulval bursting phenotypes by non-heterochronic methods. n=number of animals scored.
Abstract:
In C. elegans larvae, the execution of stage-specific developmental events is controlled by heterochronic genes, which include those encoding a set of transcription factors and the microRNAs that regulate the timing of their expression. Under adverse environmental conditions, developing larvae enter a stress-resistant, quiescent stage called 'dauer'. Dauer larvae are characterized by the arrest of all progenitor cell lineages at a stage equivalent to the end of the second larval stage (L2). If dauer larvae encounter conditions favorable for resumption of reproductive growth, they recover and complete development normally, indicating that post-dauer larvae possess mechanisms to accommodate an indefinite period of interrupted development. For cells to progress to L3 cell fate, the transcription factor Hunchback-like-1 (HBL-1) must be downregulated. Here, we describe a quiescence-induced shift in the repertoire of microRNAs that regulate HBL-1. During continuous development, HBL-1 downregulation (and consequent cell fate progression) relies chiefly on three let-7 family microRNAs, whereas after quiescence, HBL-1 is downregulated primarily by the lin-4 microRNA in combination with an altered set of let-7 family microRNAs. We propose that this shift in microRNA regulation of HBL-1 expression involves an enhancement of the activity of lin-4 and let-7 microRNAs by miRISC modulatory proteins, including NHL-2 and LIN-46. These results illustrate how the employment of alternative genetic regulatory pathways can provide for the robust progression of progenitor cell fates in the face of temporary developmental quiescence.
Legend:
Post-dauer suppression of mir-48 mir-241(nDf51); mir-84(n4037) retarded phenotypes. (A) Young adult C. elegans hermaphrodites were scored for adult alae formation (top) and col-19::GFP expression (inset and bottom). Inset shows GFP fluorescence with 10× objective. Top shows DIC image with 63× objective. Black solid lines indicate alae, wh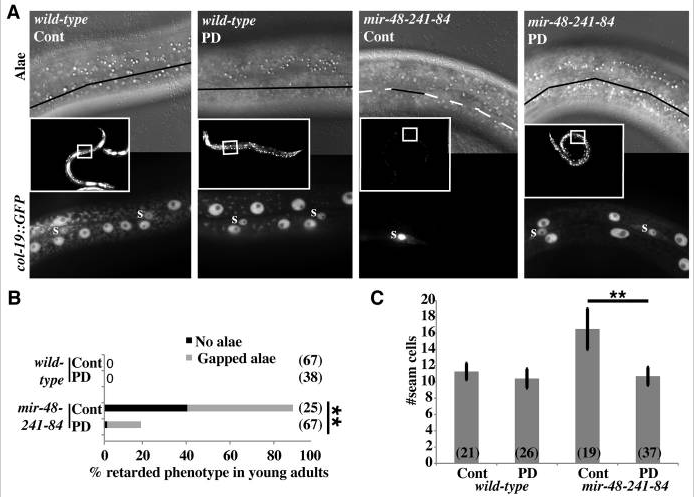 ite dashed lines indicate no alae. Bottom shows GFP fluorescence with 63× objective. GFP-positive seam cells are marked with ‘s’. Other GFP+ cells are hyp7 nuclei. (B) Percentage of young adult hermaphrodites exhibiting retarded alae defects in N2 (wild type) and VT1066 mir-48 mir-241(nDf51); mir-84(n4037). n≥25. *P<0.01, Fisher’s Exact Test. (C) Average seam cell number of L3 or post-dauer-L3 larvae of strains containing wIs51[scm::GFP]. GFP+ cells were counted between the pharynx and the rectum of each larva. n>19. Error bars indicate s.d. **P<0.01, two-tailed t-test. Cont, continuous development; PD, post-dauer.
ite dashed lines indicate no alae. Bottom shows GFP fluorescence with 63× objective. GFP-positive seam cells are marked with ‘s’. Other GFP+ cells are hyp7 nuclei. (B) Percentage of young adult hermaphrodites exhibiting retarded alae defects in N2 (wild type) and VT1066 mir-48 mir-241(nDf51); mir-84(n4037). n≥25. *P<0.01, Fisher’s Exact Test. (C) Average seam cell number of L3 or post-dauer-L3 larvae of strains containing wIs51[scm::GFP]. GFP+ cells were counted between the pharynx and the rectum of each larva. n>19. Error bars indicate s.d. **P<0.01, two-tailed t-test. Cont, continuous development; PD, post-dauer.
