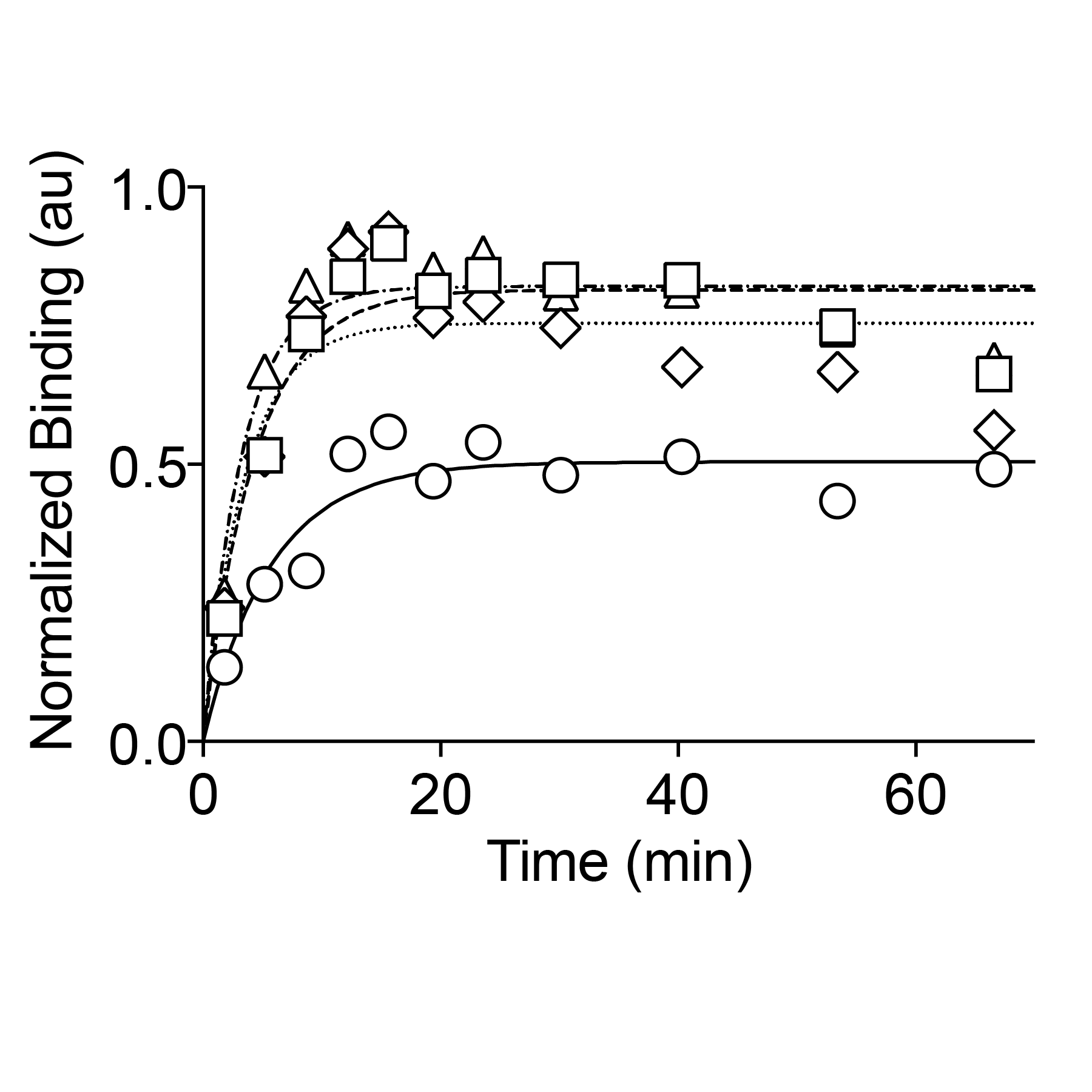
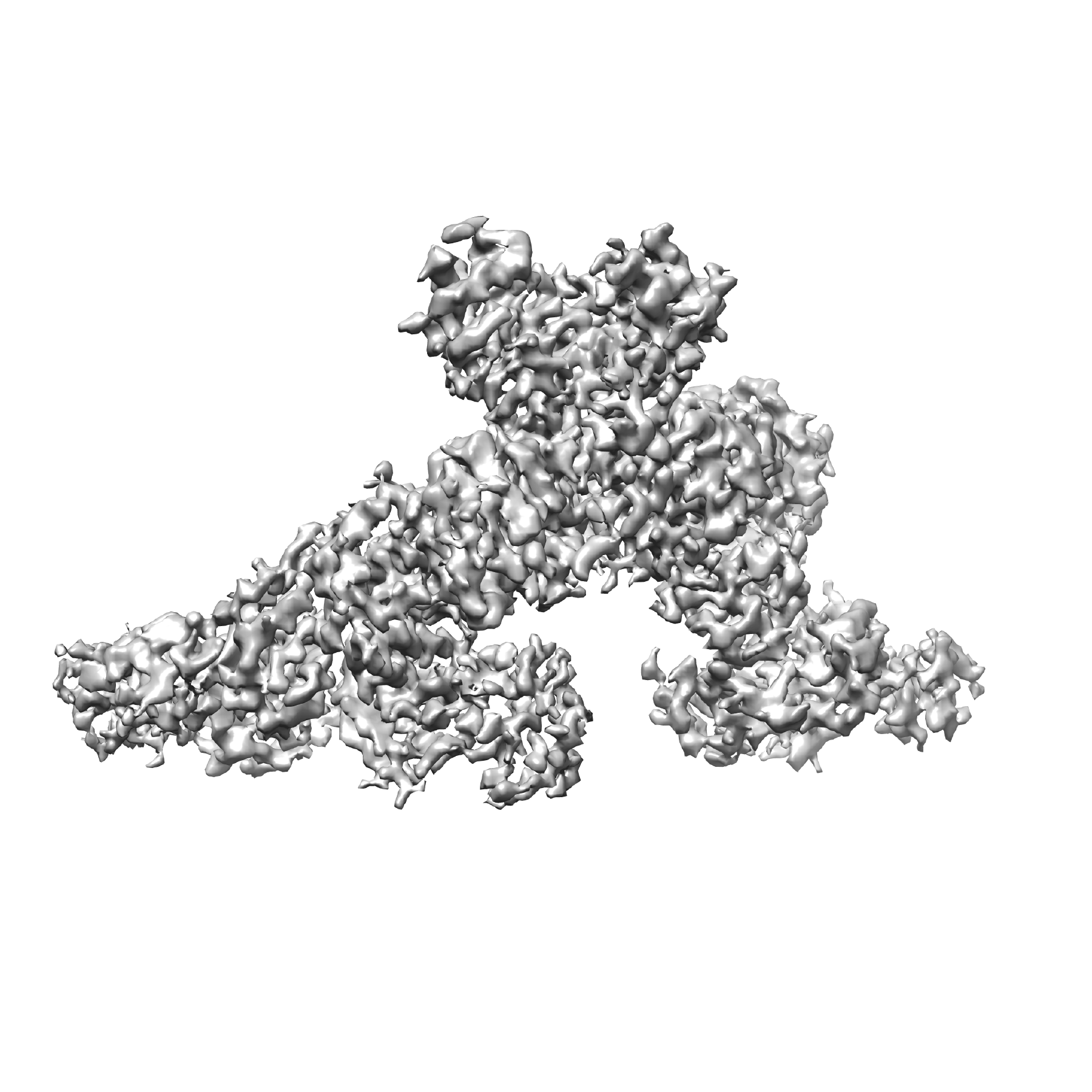
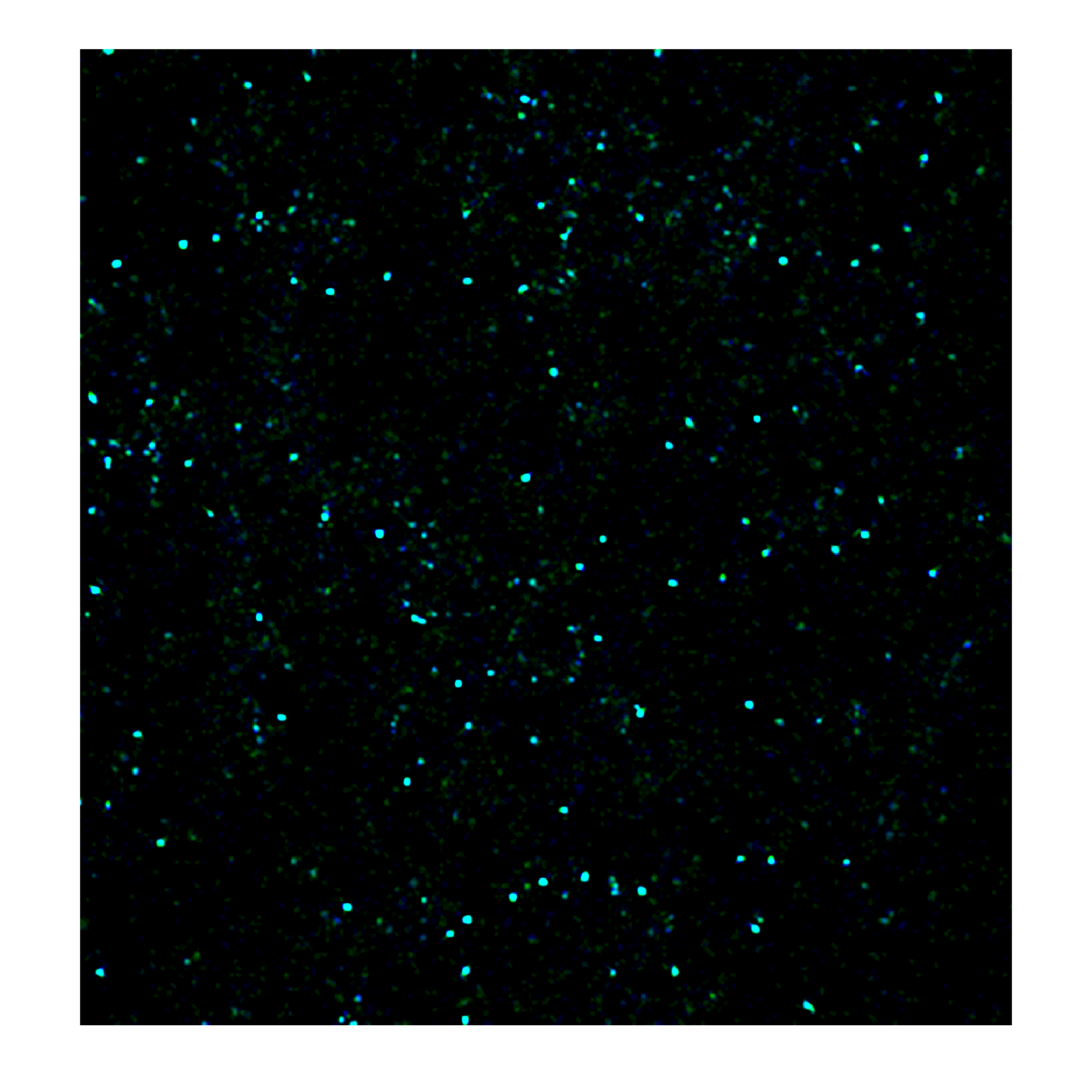
Research
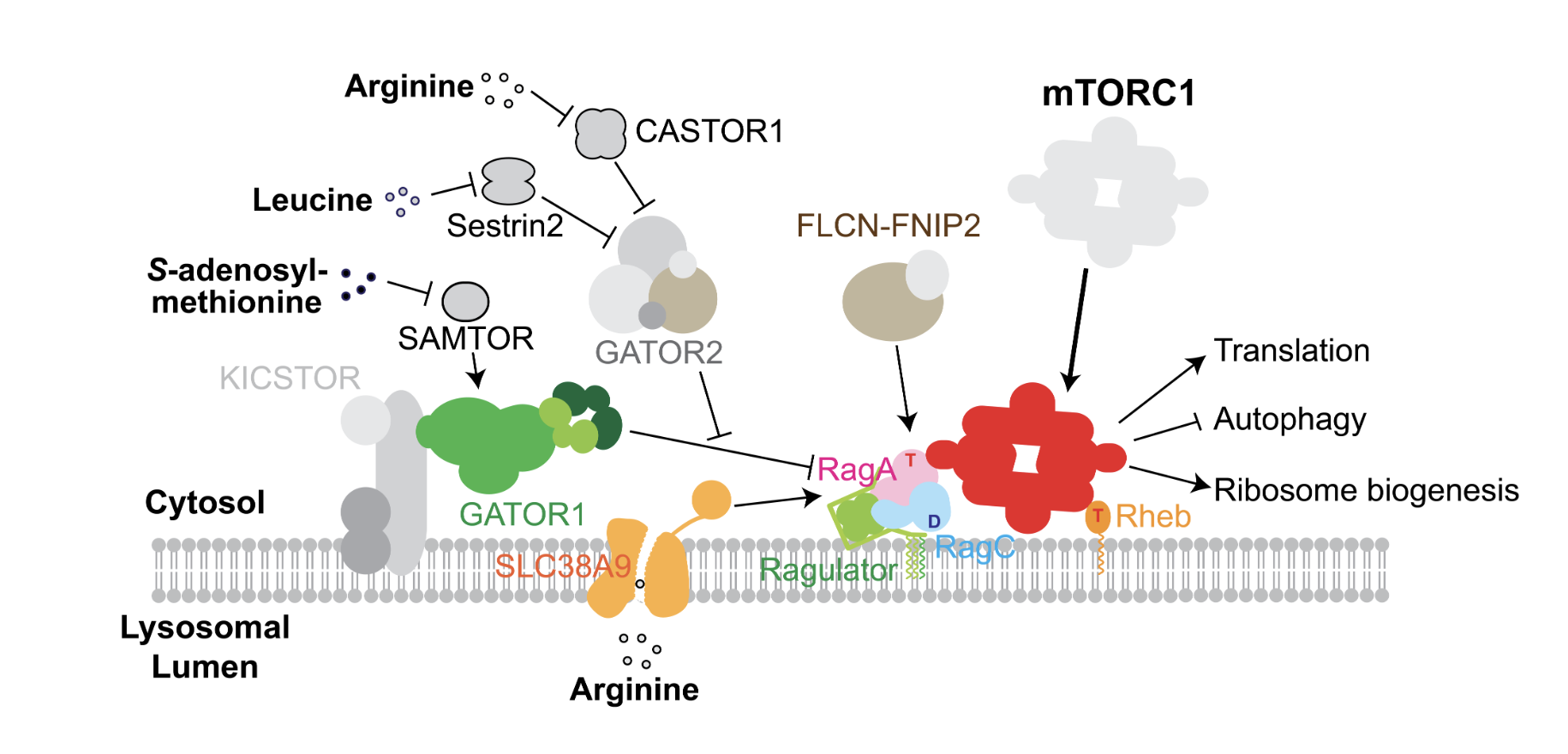
Our study focuses on nutrient sensing - in particular amino acid sensing - in eukaryotic cells. Amino acids are among the basic building blocks of all living organisms. In eukaryotic cells, a dedicated pathway, the mTORC1 pathway, senses the presence or absence of amino acids in the environment, to license cell growth. In the Shen lab, we are interested in using biochemical and biophysical tools including enzymatic kinetics, structural biology (cryo-EM), and single molecule biophysics, to investigate the mTORC1 pathway at the molecular level. We aim to gain a fundamental understanding of this biological process and how its dysfunction could lead to human disease.