The Circadian Clock: Clockwork
Our studies in the monarch butterfly have brought about a paradigm shift in how we view the evolution and function of certain clock proteins in animals (Zhu et al., 2008). What makes the monarch’s clock mechanism so unusual is the participation of two distinct CRYPTOCHROME proteins—the two CRYs of the butterfly (Zhu et al., 2005). The Drosophila-like CRY (designated CRY1) functions as a blue-light circadian photoreceptor that entrains the clock, while the vertebrate like CRY (designated CRY2) functions as the major transcriptional repressor of the clockwork transcriptional feedback loop.
The diagram below depicts a proposed monarch butterfly circadian clock mechanism. The main gear of the clock is a self-regulating transcription feedback loop. Based on the action of this feedback loop, clock protein levels oscillate in a roughly 24-hour rhythm even in the absence of light.
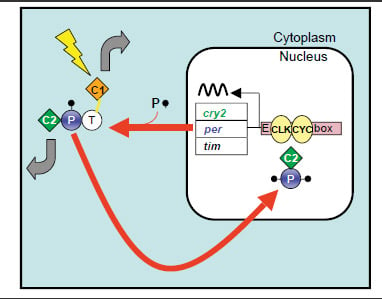
CLOCK (CLK) and CYCLE (CYC) join together in the nucleus and drive the transcription of the per, tim, and cry2 genes to make PERIOD (PER), TIMELESS (TIM), and CRYPTOCHROME 2 (CRY2). In the cytoplasm, PER, TIM and CRY2 form complexes and CRY2 is shuttled into the nucleus. There, it shuts down the transcription driven by CLK and CYC, completing the loop. Meanwhile, PER is progressively phosphorylated and likely helps translocate CRY2 into nucleus. CRYPTOCHROME 1 (CRY1) is a circadian photoreceptor which, upon light exposure (lightning bolt), causes TIM degradation to gain access to the central clock mechanism. The thick gray arrows represent output functions for CRY1 and for CRY2. Zhu H et al. PLoS Biol. 6(1): e4 (2008)
Our analysis of distinct CRY function has been further aided by a monarch cell line (DpN1 cells) that expresses a light-driven clock in which both monarch CRY1 and CRY2 function. Our continued studies will expand our knowledge of the unique properties of the CRY proteins by further defining distinct CRY mechanisms of action in lepidopterans (butterflies and moths) using molecular, biochemical, genetic, and behavioral approaches. Because of the parallel, complementary nature of circadian clock discoveries between flies and mammals, which we believe can now be extended to lepidopterans, our overriding hypothesis is that further analysis of the two families of animal CRY proteins that are represented in insects will advance our fundamental understanding of animal clock mechanisms.
Our ongoing projects include...
- Employing an RNAi screen to discover novel components of the insect CRY1 light-signaling pathway. We postulate that novel components of the CRY1-dependent circadian input pathway remain to be elucidated. The light-driven clock in DpN1 cells and a recently constructed monarch expressed sequence tag library enable the development of an unbiased, high-throughput RNAi screen to discover novel components of light-induced CRY1 signaling. This will help us understand in better detail how the molecule functions as a circadian photoreceptor. We will further analyze the mechanistic details of novel component function in DpN1 cells, and candidate components will be characterized in Drosophila strains harboring loss-of-function mutations of the fly homologs.
- Using a proteomics approach to identify novel interactors within the insect CRY2/CLK:CYC transcriptional complex. We hypothesize that yet to be discovered insect CRY2 interactive proteins are essential for its role as a major transcriptional repressor of the clockwork transcriptional feedback loop. With DpN1 cells, we will use tandem affinity purification and coimmunoprecipitation to identify additional CRY2-, CLOCK-, and CYCLE-interacting proteins. Finding novel interactors may help define the transcriptional repressive role of CRY not only in insects, but also in mammals, in which the mechanism of CRY repression is not well understood.