Electric signals play important roles in diseases ranging from cardiac arrhythmias to diabetes.
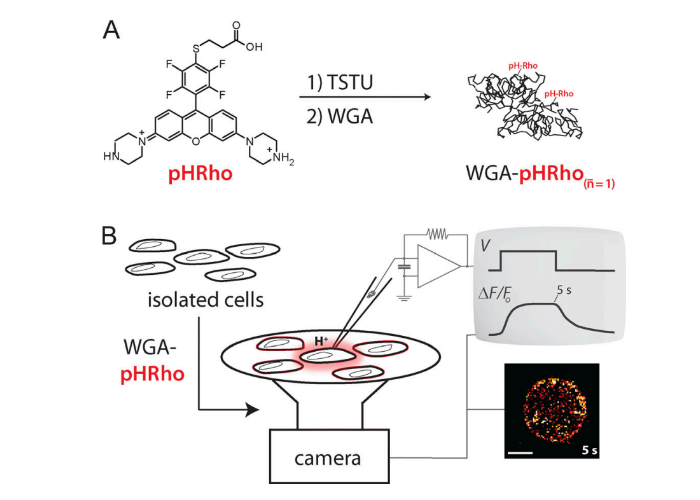 Wheat germ agglutinin-conjugated fluorescent pH sensors for visualizing proton fluxes.
Wheat germ agglutinin-conjugated fluorescent pH sensors for visualizing proton fluxes.
Zhang L, Zhang M, Bellve K, Fogarty KE, Castro MA, Brauchi S, Kobertz WR.
J Gen Physiol. 2020 Jun 1;152(6):e201912498. doi: 10.1085/jgp.201912498. PMID: 31978216; PMCID: PMC7266149.
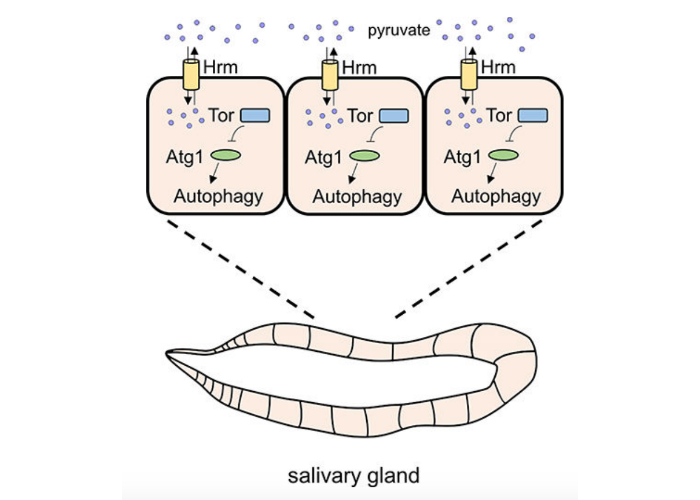 The Proton-Coupled Monocarboxylate Transporter Hermes Is Necessary for Autophagy during Cell Death.
The Proton-Coupled Monocarboxylate Transporter Hermes Is Necessary for Autophagy during Cell Death.
Velentzas PD, Zhang L, Das G, Chang TK, Nelson C, Kobertz WR, Baehrecke EH.
Dev Cell. 2018 Nov 5;47(3):281-293.e4. doi: 10.1016/j.devcel.2018.09.015. Epub 2018 Oct 11. PMID: 30318245; PMCID: PMC6219939.
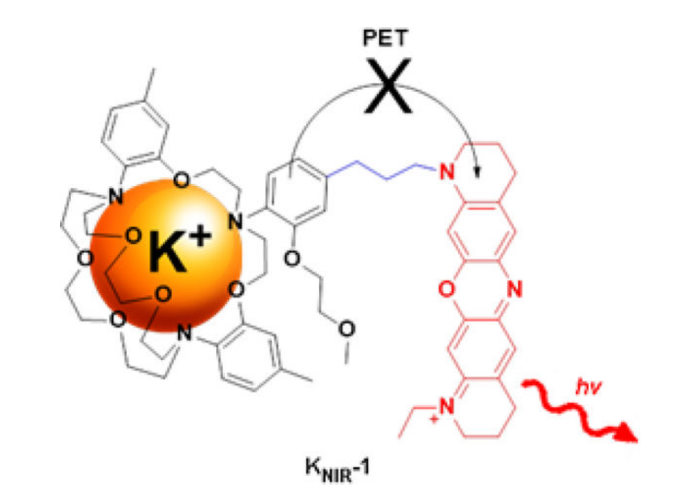 Palladium-Mediated Synthesis of a Near-Infrared Fluorescent K+ Sensor.
Palladium-Mediated Synthesis of a Near-Infrared Fluorescent K+ Sensor.
Bandara HMD, Hua Z, Zhang M, Pauff SM, Miller SC, Davie EAC, Kobertz WR.
J Org Chem. 2017 Aug 4;82(15):8199-8205. doi: 10.1021/acs.joc.7b00845. Epub 2017 Jul 14. PMID: 28664732; PMCID: PMC5715468.



