Research
The Genetics of Immune Activation and Regulation in Intestinal Epithelial Cells Intestinal epithelial cells are the primary interface with commensal microorganisms, potential pathogens and ingested toxins, and function to maintain mucosal homeostasis. Immune regulation is particularly complex in intestinal epithelial cells, since these cells must perform the dual roles of protecting the host from potential pathogens while preventing exaggerated inflammatory responses that can cause collateral injury to the host. What are the mechanisms that ensure immune homeostasis in the intestinal epithelium? How do these cells distinguish between beneficial microbes and invasive pathogens, and mount targeted immune responses only in the context of infection?
Intestinal epithelial cells are the primary interface with commensal microorganisms, potential pathogens and ingested toxins, and function to maintain mucosal homeostasis. Immune regulation is particularly complex in intestinal epithelial cells, since these cells must perform the dual roles of protecting the host from potential pathogens while preventing exaggerated inflammatory responses that can cause collateral injury to the host. What are the mechanisms that ensure immune homeostasis in the intestinal epithelium? How do these cells distinguish between beneficial microbes and invasive pathogens, and mount targeted immune responses only in the context of infection?
Our laboratory approaches these problems by focusing on conserved mechanisms of immune activation. The major hypothesis of our work is that the mechanisms that act in intestinal epithelial cells to distinguish pathogens from commensals have evolutionarily ancient origins, the fundamental principals of which can be elucidated using a simple model host. In nature, the genetically tractable soil nematode Caenorhabditis elegans consumes microorganisms for food and thus, their evolution has been shaped by interactions with microbial pathogens and ingested toxins. We therefore take advantage of the powerful molecular and genetic tools that are available in C. elegans and a human bacterial pathogen Pseudomonas aeruginosa that is also amenable to genetic manipulation, to define conserved mechanisms of pathogen detection and immune regulation.
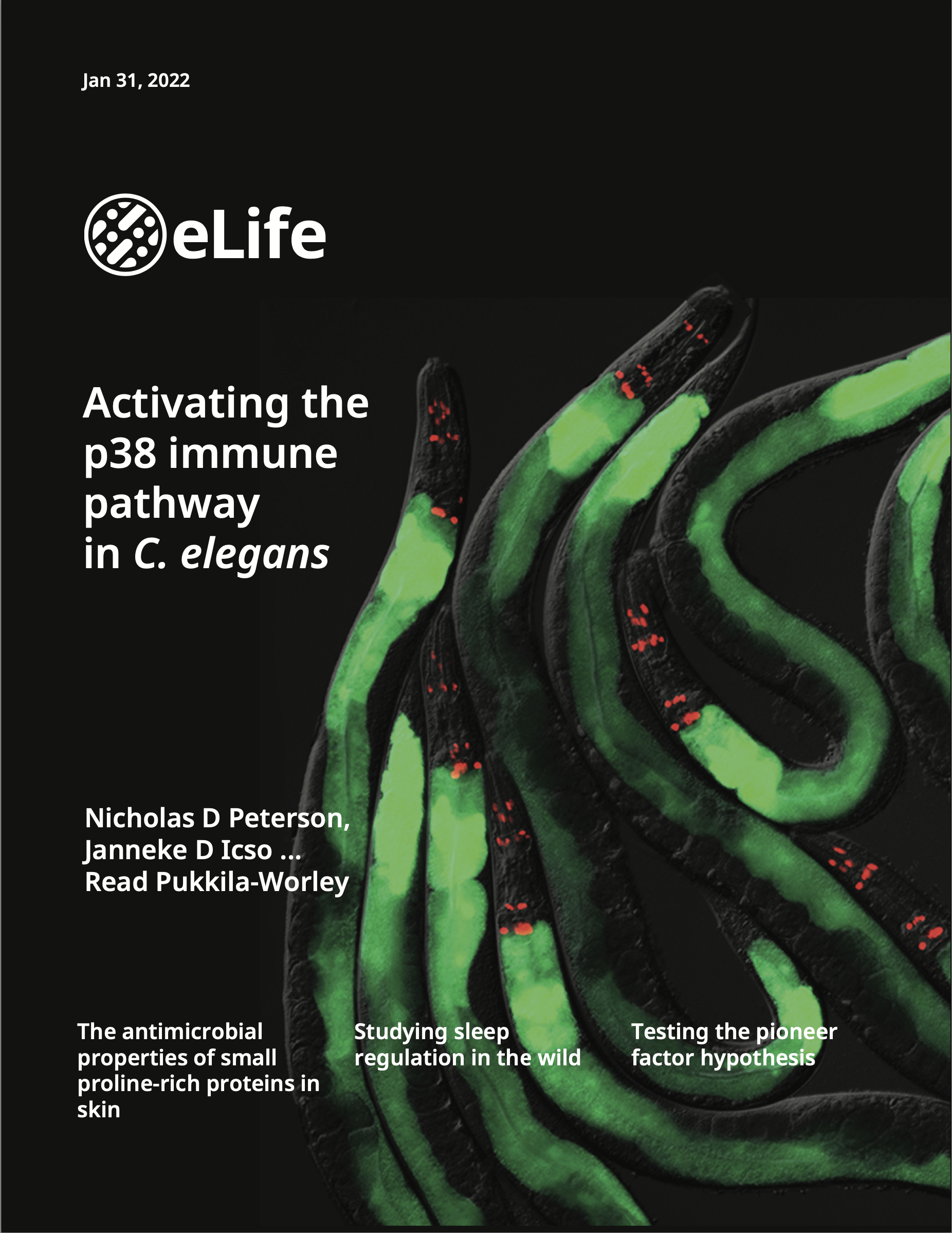
These genetic, cell biological and molecular biological studies in C. elegans and Pseudomonas aeruginosa have: characterized a non-canonical pattern recognition system that intercepts pathogen-derived signals of growth and virulence to assess the relative threat of virulent bacteria (Immunity 2023); revealed that both pathogen infection and cholesterol deficiency activate the p38 immune signaling pathway in the intestine by inducing oligomerization and a phase transition of a key immune signaling regulator (eLife 2022); characterized an unexpected connection between neuronal development and innate immunity, which revealed that anti-pathogen defenses in the intestine are developmentally programmed (Cell Reports 2020); defined a mechanism that allows intestinal cells to counteract the pleiotropic consequences of aberrant immune activation and discovered a new immunometabolic phenotype of pathogen infection (PNAS 2019); uncovered a novel connection between lipid metabolism and host susceptibility to bacterial infection (PLOS Pathogens 2019); characterized a new mechanism of pathogen sensing in intestinal epithelial cells (PLOS Genetics 2019); and demonstrated that innate immune control in the intestine is required for the proper development of a metazoan host (G3 2016).We also received a Notice of Allowance on a U.S. Patent (Application 16/069,399, filed January 11, 2016) for our discovery that genetic inactivation of a cell surface receptor in nematodes triggers toxic immune hyperactivation, which we propose can be targeted to generate new anti-helminthic medications.
These studies have led us to focus on three principal areas of current interest: 1) Immune pathway activation in IECs by nuclear hormone receptors. Our data suggest that the expansion of the nuclear hormone family in C. elegans has been fueled, at least in part, by the roles of these proteins in the activation of host defense responses. 2) p38 MAP kinase innate immune signaling. We take cell and molecular biological approaches to characterize new mechanisms of regulation and function of this evolutionarily ancient immune regulatory pathway. 3) Lipid metabolism and immune activation. We are defining the genetic links between lipid metabolism and the activation of protective immune defenses.
Together, these efforts are uncovering fundamental principles of immune homeostasis involving conserved immune regulators that we are applying to mammalian biology using cell culture and mouse genetics.