The Substrate Envelope
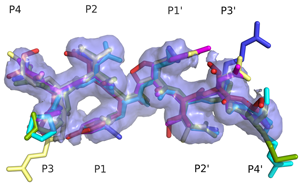
Specificity Determinants of Natural Substrate RecognitionHIV-1 protease processes the viral Gag and Gag-Pro-Pol polyproteins at twelve cleavage sites. These cleavage sites share no obvious sequence homology. However, when bound to the active site of the protease, these natural substrates adopt a conserved consensus volume. We define this consensus volume as the substrate envelope. According to our substrate envelope hypothesis, the substrate envelope is the recognition motif for HIV-1 protease. (Prabu-Jeyabalan et. al., 2002) |
Superimposed protease-bound natural substrates reveal a consensus volume; the substrate envelope. |
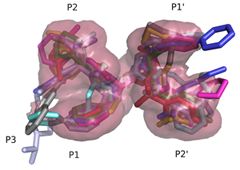
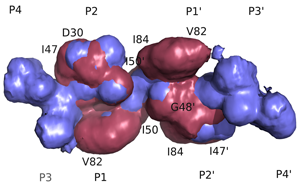
Structural Basis of Drug ResistanceFDA-approved HIV-1 protease inhibitors are all chemically different; yet share a fairly similar three-dimensional shape and electrostatic character. They also adopt a conserved consensus volume in the binding site of the enzyme. We define this consensus volume as the inhibitor envelope. The Inhibitor envelope, superimposed onto the substrate envelope, protrudes beyond the substrate envelope and makes contacts with certain protease residues. These residues correspond to the previously known primary drug resistance mutation sites. These sites are presumably more important for inhibitor binding versus substrate processing, hence they mutate under the selective pressure of the drug therapy yielding drug-resistant viral strains. (King et. al., 2004)
|
Superimposed protease-bound inhibitors reveal a consensus inhibitor volume; the inhibitor envelope.
Inhibitors contact the drug resistance mutation sites in the protease outside the substrate envelope |
New Paradigm in Drug Design: Avoid resistance in drug design process
The inhibitors, when designed to fit within the substrate envelope, are less likely to elicit drug-resistance mutations.
Using Substrate Dynamics to define the Dynamic Substrate Envelope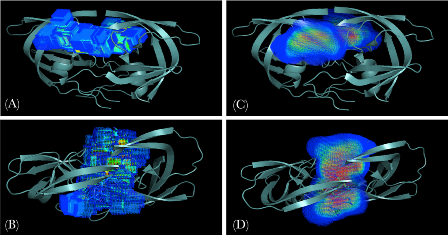
Protein recognition is inherently a dynamic event because proteins adopt an ensemble of different conformations around their native states. Consideration of this flexibility around the native state is essential to arriving at a better understanding of the specificity determinants of substrate recognition. Introducing molecular dynamics into the substrate envelope allows a more realistic evaluation of the substrate envelope hypothesis. In addition, the dynamic substrate envelope is a probability distribution of the consensus substrate volume. This probability distribution function can easily be incorporated as a constraint into the scoring functions in computer-aided drug design algorithms. (Ozen et. al., 2011)