Molecular Basis for Resistance
Molecular Mechanisms of Drug Resistance – Viral proteases as model systems
Viruses can evolve very quickly and acquire mutations that cause drug resistance. We take an integrated multi-disciplinary approach to understanding resistance combining the strengths of: protein crystallography (solved over 160 crystal structures); isothermal titration calorimetry; computational structural biology including: extensive molecular dynamics simulations, homology modeling and docking; sequence analysis; organic synthesis (synthesized over 200 compounds) and, through collaboration, virology and NMR.
Drug resistance at the molecular level is a subtle change in the balance of recognition events between the relative affinity of the enzyme to bind inhibitors and its ability to process substrates. HIV-1 evolves extremely quickly, due to error prone replication and host APOBEC3s promoting extreme viral diversity. Thus HIV is an ideal model system to mine the mechanisms of drug resistance. HIV-1 protease, with the plethora of available experimental data, has been our ideal prototype for drug design against quickly evolving targets, allowing us to elucidate the fundamentals of drug resistance. Enriching and exploiting this rich data, we are developing new strategies such as incorporation of the substrate envelope constraint to drug design to counter drug resistance. These strategies have proven applicable to other targets including HCV protease and we are extending ideas to other viral proteases (Zika, Dengue) and disease targets (influenza, cancer).
Manuscript Highlights
 |
|
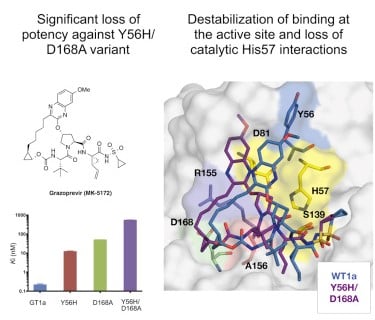
Molecular Mechanism of Resistance in a Clinically Significant Double-Mutant Variant of HCV NS3/4A Protease. |
 |
|
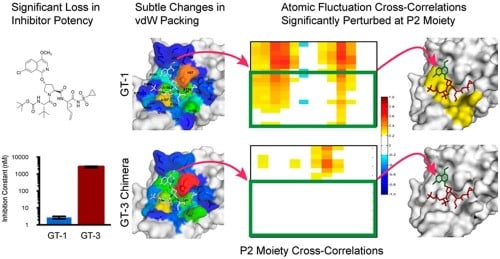
Molecular and Dynamic Mechanism Underlying Drug Resistance in Genotype 3 Hepatitis C NS3/4A Protease. |

