How important is maternal RNA regulation to reproduction?
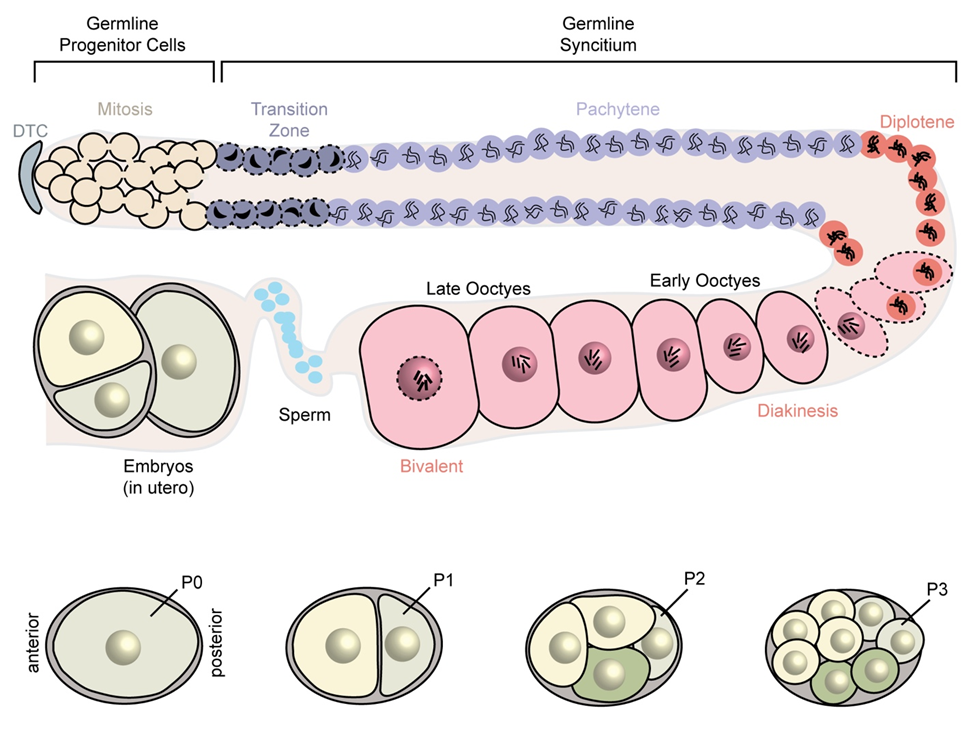
My lab uses Caenorhabditis elegans to study maternal RNA regulation. Forward genetic screens identified numerous genes that when mutated give rise to maternal effect lethal or sterile phenotypes caused by failures in embryogenesis and/or gametogenesis. Many encode RNA-binding proteins and/or their target mRNAs. Our niche over the past twenty years has been to map germline RBP specificity using quantitative methods and assess the functional relevance in vivo using integrated transgenic fluorescent reporters. As a result, we have built a wide array of tools to study nematode RBP structure and function. But these tools cannot tell us which regulatory events are most important to reproduction and under what circumstances. Of the 2,152,744 “variations”—SNPs, indels, alleles, etc.—listed in WormBase, only a small fraction (57,219: 2.66%) are found within 3´UTRs. We do not know how many of these have a functional consequence. WormBase lists 8416 “classical alleles” identified through forward genetic screens. Only 33 (0.39%) fall within 3´UTRs. Yet reporter studies show that the 3´UTRs of maternal mRNAs are the primary determinants of their expression pattern. But which, if any, matter to reproduction? And how do they work?
Until recently, we lacked the tools to make site directed mutations at endogenous loci in C. elegans. Advances in CRISPR-Cas9 genome editing have eliminated this roadblock. Genome editing enables targeted gene disruptions and replacements, as well as insertion of fluorescent proteins, epitope tags, and degrons all at the endogenous locus. We have used this method to make deletions and targeted substitutions within the 3´UTR of critical maternal effect genes to determine how important RNA regulation is to reproduction.
Recent Publications:
Brown, H.E., Varderesian, H.V., Lam, O., Keane, S., and Ryder, S.P., (2025) The mex-3 3’UTR is essential for reproduction during temperature stress. Development. 152 (17): dev204740
Antkowiak, K.†, Coskun, P.†, Noronha, S.†, Tavelle, D., Massi, F., and Ryder, S.P., (2024) A nematode model to evaluate microdeletion phenotype expression. G3 Genes|Genomes|Genetics.14 (2): jkad258, https://doi.org/10.1093/g3journal/jkad258
Albarqi, M.M.Y. and Ryder S.P. (2021) The endogenous mex-3 3’UTR is required for germline repression and contributes to optimal fecundity in C. elegans, PLOS Genet. 2021 Aug 23;17(8):e1009775. doi: 10.1371/journal.pgen.1009775.

