Protein Interactions via Disordered Peptide Sequences
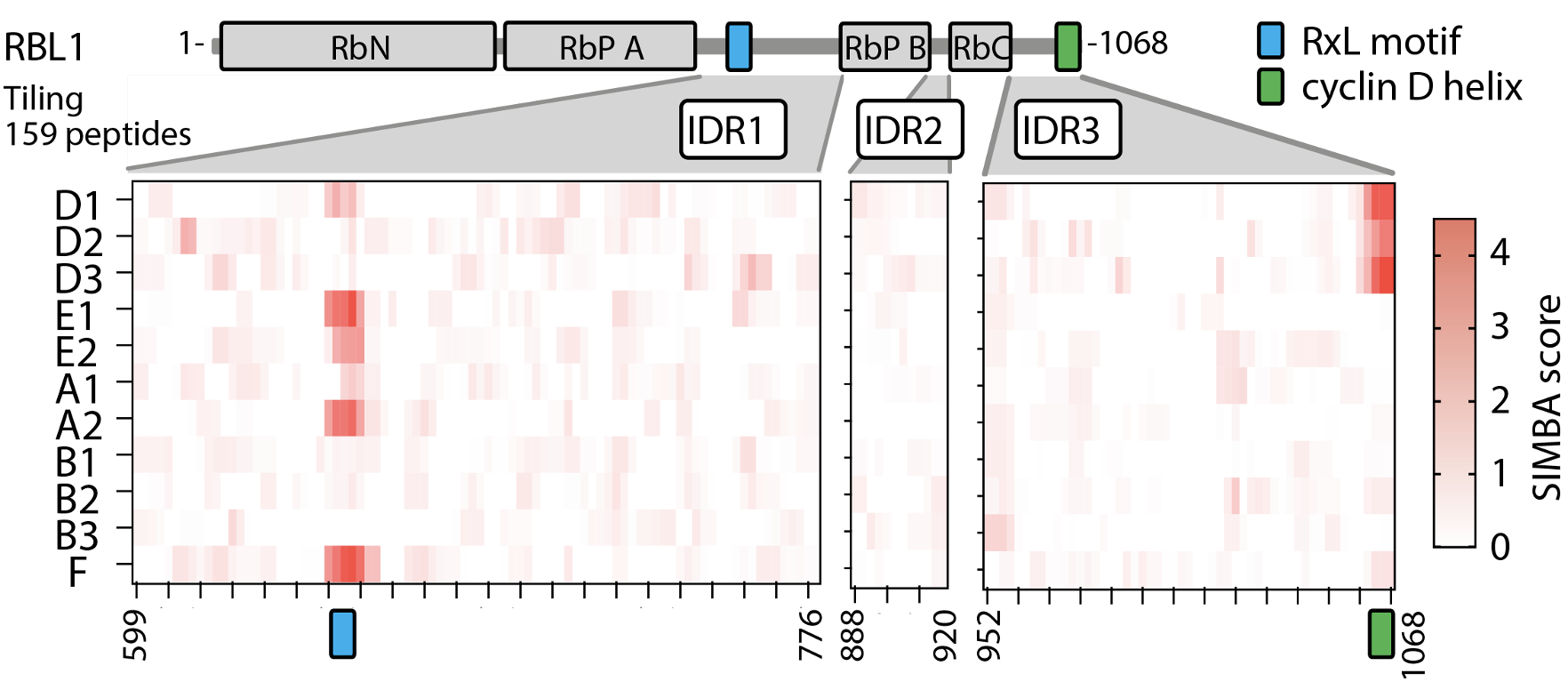
A wide variety of cellular functions are controlled by weak, transient protein-protein interactions in which a globular folded domain in one protein binds to a short linear motif (SLiM) in a disordered region of its partner protein. The human proteome contains hundreds of SLiM-binding domains, yet in most cases the key sequence features of SLiMs that govern their recognition are poorly defined. Because cells use variations in SLiM binding affinity to tune their physiological effects, it is critical to decipher how affinity is dictated by sequence. We have developed a novel functional assay for measuring SLiM binding, called Systematic Intracellular Motif Binding Analysis (SIMBA). This method allows us to quantify the binding strength of a peptide-binding domain to thousands of different peptide motifs simultaneously. We are using this tool and others to investigate the role of SLiMs in intracellular regulatory pathways, including those governing cell division. Each step of cell division is coordinated by cyclin-dependent kinases (CDKs) that phosphorylate hundreds of proteins in a temporally resolved manner. SLiMs help control this orderly progression of regulatory steps. Namely, the cyclin subunit can regulate CDK specificity by binding SLiM "docking" peptides in CDK substrates. We are actively investigating the contributions of SLiMs to cell cycle control as well as several other important regulatory networks.
Relevant Publications
Bandyopadhyay S, Bhaduri S, Örd M, Davey NE, Loog M, Pryciak PM. Comprehensive Analysis of G1 Cyclin Docking Motif Sequences that Control CDK Regulatory Potency In Vivo. Curr Biol. 2020 Nov 16;30(22):4454-4466.e5. doi: 10.1016/j.cub.2020.08.099. Epub 2020 Sep 24. PMID: 32976810; PMCID: PMC8009629.
Subbanna MS, Winters MJ, Örd M, Davey NE, Pryciak PM. A quantitative intracellular peptide binding assay reveals recognition determinants and context dependence of short linear motifs. bioRxiv [Preprint]. 2024 Nov 1:2024.10.30.621084. doi: 10.1101/2024.10.30.621084. PMID: 39553988; PMCID: PMC11565833.
