 Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow
Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow
2016 doi.org/10.1038/nbt.3526 CRISPRainbow.
Hanhui Ma, Li-Chun Tu, Ardalan Naseri, Shaojie Zhang, David Grunwald and Thoru Pederson
This innovation expanded the range of colors that can be used to simultaneously used track the movements of as many as six different genomic sites in a single, living cell.
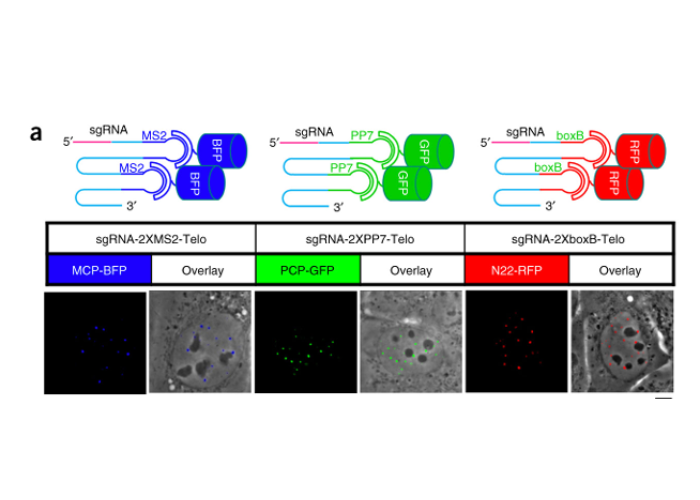
CRISPR Sirius: RNA scaffolds for signal amplification in genome imaging
2018 doi.org/10.1038/s41592-018-0174-0
Hanhui Ma, Li-Chun Tu, Ardalan Naseri, Yu-Chieh Chung, David Grunwald, Shoajie Zhang and Thoru Pederson.
In this method the guide RNAs were modified so as to bind a larger number of fluorescent proteins than before, enhancing the detection sensitivity of labeled genomic loci.
Simultaneous epigenetic perturbation and genome imaging reveal distinct roles of H3K9me3 in chromatin architecture and transcription.
2020 doi.org/10.1186/s13059-020-02201-1
Ying Feng, Yao Wang, Xiangnan Wang, Xiaohui He, Cheng Yang, Ardalan Naseri, Thoru Pederson, Jing Zheng, Shaojie Zhang, Xiao Xiao, Wei Xie and Hanhui Ma.
By attaching to the dCas9 a protein element that induces a specific chemical modification of chromatin, effects on transcription vs. genome organization could be differentiated.



