Why study membrane trafficking?
We are interested in understanding the mechanistic basis for regulation of the spatial and temporal specificity of membrane trafficking. Many questions remain to be answered regarding how cargos arrive at the correct location. For example, what marks the site of vesicle fusion on the target membrane and what checks to make sure that the correct vesicle docks at the right place? How are the membrane fusion proteins (SNAREs) regulated to ensure that the wrong vesicle does not fuse? Our aim is to answer questions such as these through a multifaceted approach that combines biochemical, structural and biophysical techniques with yeast and mammalian genetics, microscopy and cell biological methods. We are investigating proteins that regulate membrane trafficking in the yeast Saccharomyces cerevisiae, in mice, and in human cell lines. Because these proteins are highly conserved, these studies will advance our understanding of how membrane trafficking is regulated in all eukaryotic cells. Our research is mainly focused in three areas:
Control of Exocytosis by the Exocyst Complex and Sec1
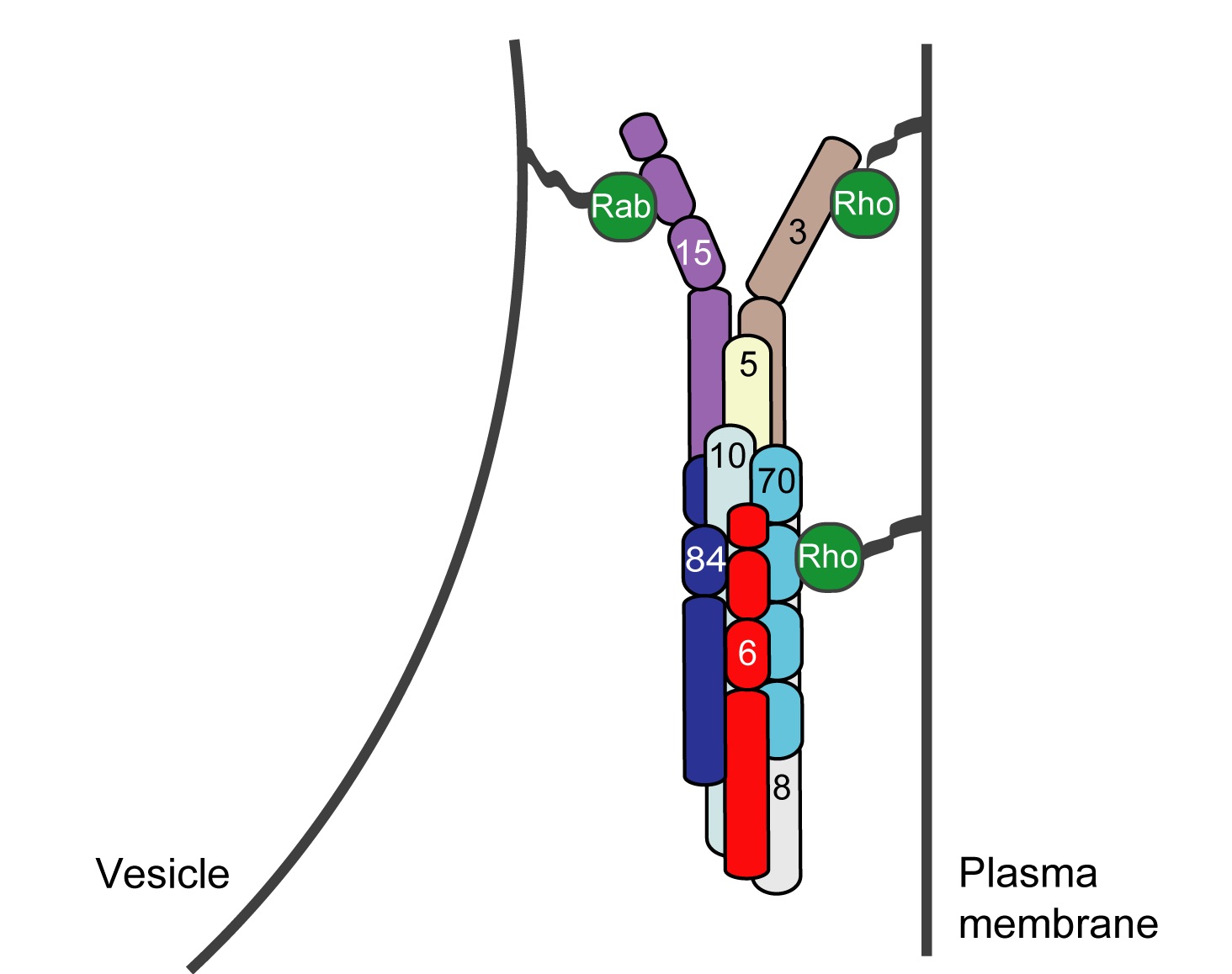
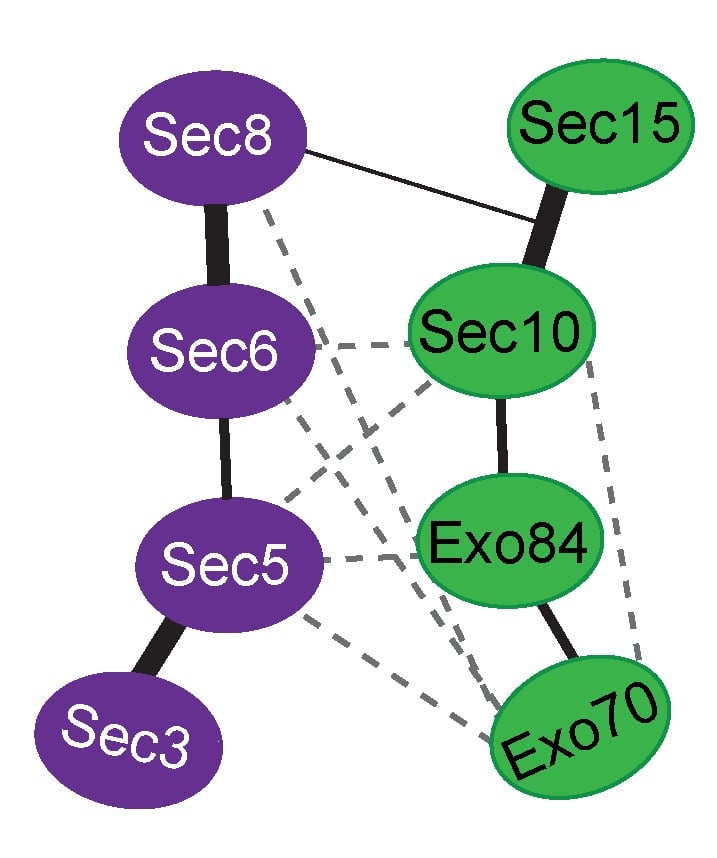
Exocytosis is the regulated fusion of secretory vesicles at the plasma membrane, to promote cell growth, division, and intercellular communication. We study how SNARE-mediated fusion of transport vesicles at the plasma membrane is temporally and spatially controlled by the exocyst, a large multisubunit tethering complex, in collaboration with the SNARE-regulator Sec1. To answer these questions, we use biochemical and structural methods, combined with examination of specific mutant phenotypes in yeast using microscopy and secretion assays. We discovered the first direct interaction between the exocyst and SNARE proteins, and recently mapped interactions between them. Also, we identified residues on the subunit Sec6 that are essential for proper localization of exocyst to sites of exocytosis. Furthermore, with several excellent collaborators, we examined direct interactions of exocyst with the myosin motor that carries secretory vesicles. We determined the crystal structure of the exocyst subunit Sec6, and showed that it is structurally homologous to other exocyst subunits (with <10% sequence identity), and members of other tethering complexes. Furthermore, we modeled the structures of the other subunits, and predicted that they also have similar helical bundle structures. Our recent work demonstrates our groundbreaking purification method for intact yeast exocyst complexes, our genetic and biochemical dissection of the architecture of the exocyst, and the first view of the overall structure of the complex using negative stain EM (Heider et al., 2016; in collaboration with Mike Rout’s lab at Rockefeller http://lab.rockefeller.edu/rout/ and Adam Frost’s lab at UCSF http://frostlab.org/). Our goal is to answer many essential questions about how the exocyst functions in the cell--what does it look like? How does it bind to its partners, including the Rab GTPase Sec4? How is it localized? How does it tether vesicles? How does it function with Sec1 to control the SNARE fusion machinery? Current research focuses on higher resolution structural studies as well as development of two in vitro TIRF-based single molecule assays to directly examine the roles of exocyst in a) tethering (with Jeff Gelles at Brandeis Univ; http://blogs.brandeis.edu/engineshop/) and b) SNARE-mediated fusion of vesicles (with Tae-Young Yoon at Seoul National Univ, South Korea; http://newbiosci.snu.ac.kr/yoonlab/en).
Mutations in VPS45 that Lead to Severe Congenital Neutropenia
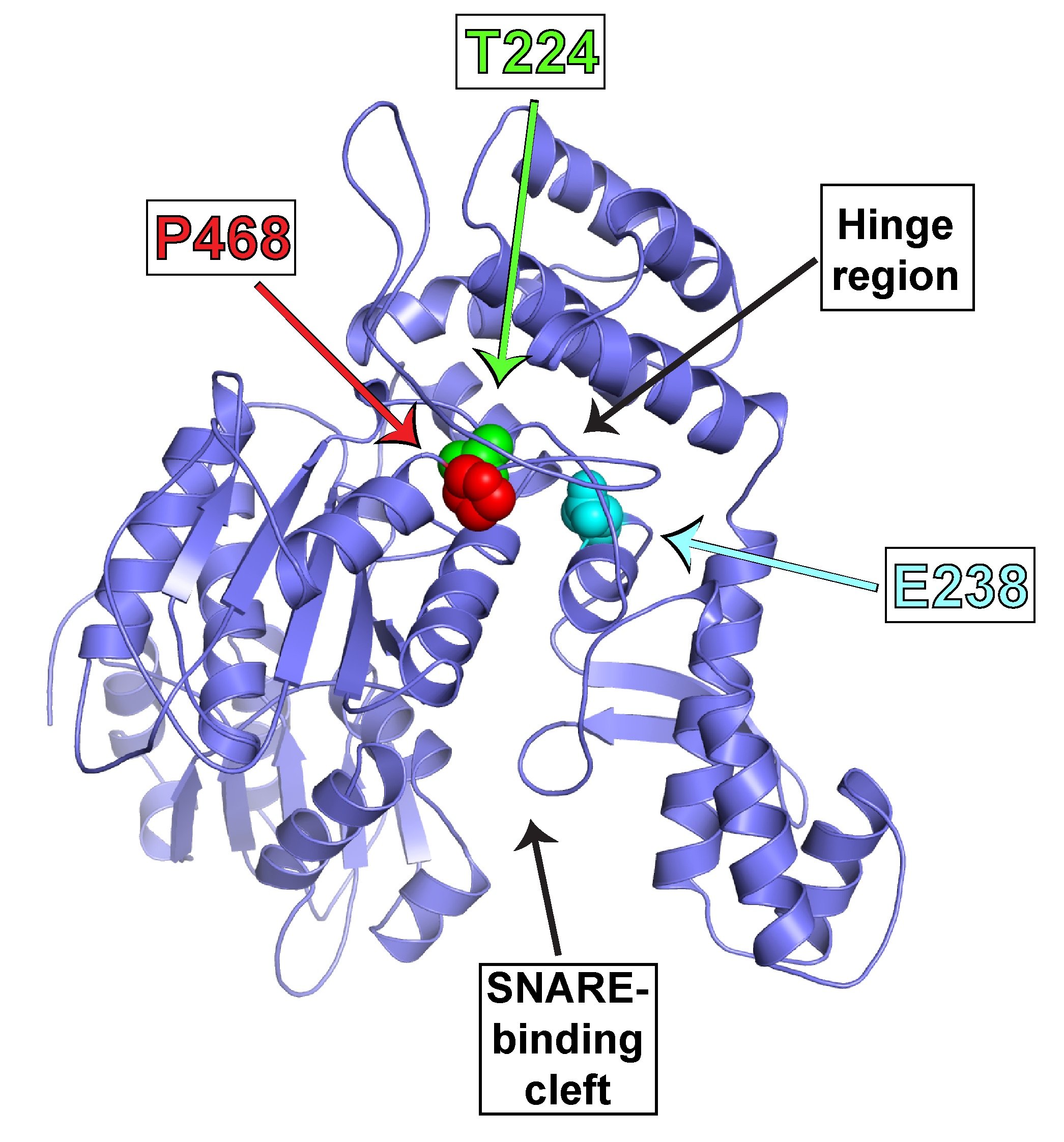
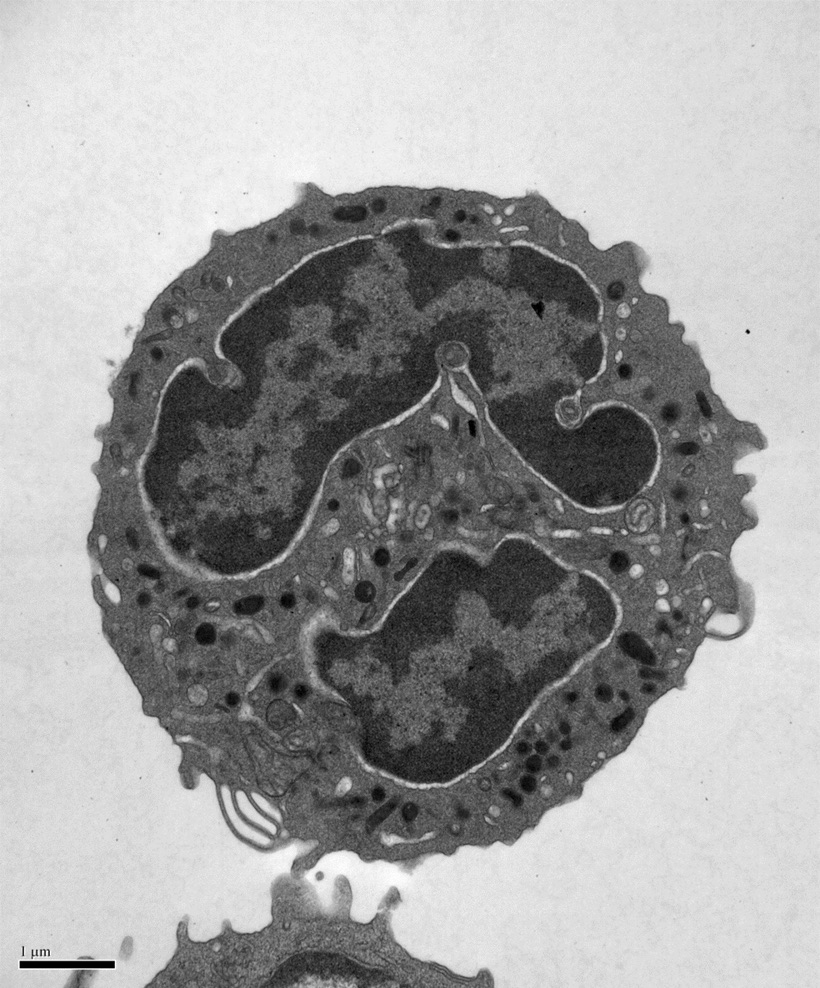
During our studies of SNARE regulation, we also investigated the endosomal Sec1/Munc18 homolog, VPS45 and dissected how it regulates SNARE complex assembly and membrane fusion. We are now applying our biochemical insights to understanding the human VPS45 and how dysfunctional point mutations in VPS45 lead to severe congenital neutropenia, bone marrow defects, and myelofibrosis in children. We are using a variety of techniques, in collaboration with Peter Newburger’s lab at UMass Chan (https://profiles.umassmed.edu/display/132970) to elucidate the mechanism behind this phenotype. These include biochemistry and structural biology methods to probe the effect of the mutations on the structure of VPS45 and its ability to interact with known binding partners; in complementary studies, cell biological techniques and fluorescence microscopy are used to understand the effect of the VPS45 mutations on cellular trafficking and organelle biogenesis. Mutant cells and mouse models are being developed using CRISPR/Cas9 technologies.

