RNA Sequencing/Bioinformatics core
Objectives
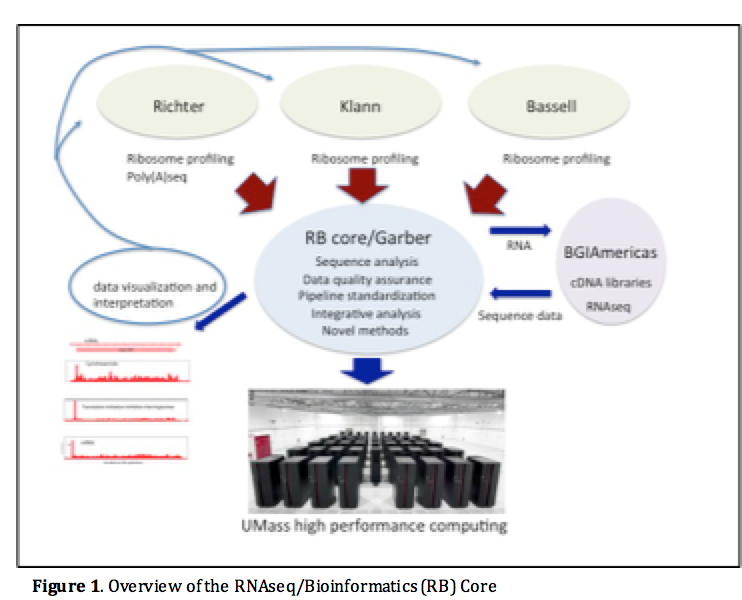
The computational core will support the efforts of all projects in this Center, its specific goals are (Figure 1):
- Provide storage, backup, access and visualization of RNA sequence datasets generated. The Core will serve as a clearinghouse for all sequence data allowing all projects to access data generated by the other projects.
- Changes in RTT are associated with Fragile X syndrome (FXS) phenotypes; this RNAseq/bioinformatics (RB) Core will thus Develop, implement, and apply RTT/ribosome profiling algorithms and methods and provide a robust analysis pipeline to quantify ribosomal run-off (ribosome transit time, RTT) followed by ribosome profiling. .
- Establish a computational pipeline for accurate measurement of polyA length using polyASeq data.
- Conduct custom integrative analyses for merged datasets to create synergies between component projects
- Coordinate cDNA library synthesis and sequencing and therefore ensure common sequencing standards (protocol, depth, read length, etc.).
- Advise in the design of genome-wide experimental design.Dr. Garber is an expert in sequence analysis and has worked with every sequencing platform since 20051-7. He was a key member of the human genome project at the Broad Institute, where he worked on chromosomal structure2,8 and in applying next generation sequencing methods to genome finishing1. Dr. Garber is a well-known expert in RNA-Seq analysis, where his review on computational methods for analysis of these data is still one of the most highly cited5. Dr. Garber has developed and maintains sequence analysis tools that will be fundamental to the analysis we propose, such as Scripture4 and Bam2X (http://bam2xwiki.appspot.com/). Since his arrival at UMASS Dr. Garber and Dr. Richter have established a close collaboration to develop sequencing based protocols and analytical approaches to study translational processes.
Manuel Garber PhD, Core Director
 Dr. Garber is an expert in sequence analysis and has worked with every sequencing platform since 20051-7. He was a key member of the human genome project at the Broad Institute, where he worked on chromosomal structure2,8 and in applying next generation sequencing methods to genome finishing1. Dr. Garber is a well-known expert in RNA-Seq analysis, where his review on computational methods for analysis of these data is still one of the most highly cited5. Dr. Garber has developed and maintains sequence analysis tools that will be fundamental to the analysis we propose, such as Scripture4 and Bam2X (http://bam2xwiki.appspot.com/). Since his arrival at UMASS Dr. Garber and Dr. Richter have established a close collaboration to develop sequencing based protocols and analytical approaches to study translational processes.
Dr. Garber is an expert in sequence analysis and has worked with every sequencing platform since 20051-7. He was a key member of the human genome project at the Broad Institute, where he worked on chromosomal structure2,8 and in applying next generation sequencing methods to genome finishing1. Dr. Garber is a well-known expert in RNA-Seq analysis, where his review on computational methods for analysis of these data is still one of the most highly cited5. Dr. Garber has developed and maintains sequence analysis tools that will be fundamental to the analysis we propose, such as Scripture4 and Bam2X (http://bam2xwiki.appspot.com/). Since his arrival at UMASS Dr. Garber and Dr. Richter have established a close collaboration to develop sequencing based protocols and analytical approaches to study translational processes.
