APOBEC3 proteins: Potential host-encoded antiviral targets
 |
|
Residues conserved among APOBEC3G and 3A are shown in silver. |
Cytidine deaminases edit nucleic acids by deaminating cytidines to uridines and are evolutionarily conserved. The biochemical simplicity of cytidine deamination mechanism has enabled AID/APOBEC proteins to evolve into powerful polynucleotide mutators. AID and the APOBEC3 proteins play key roles as DNA mutators in humoral immunity during antigen-driven antibody diversification as well as in innate antiviral and antibacterial defense. APOBEC3G/F/H exert antiretroviral effect by being incorporated into viral particles and introducing point mutations into the viral genome, thus disabling the production of functional viral proteins upon subsequent infection.
 |
|
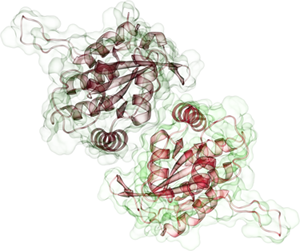
APOBEC3G Dimer |
APOBEC3A, an extremely potent DNA mutator, degrades foreign DNA in phagocytic cells and have recently been shown to play a role cancer development. Furthermore, AID/APOBEC3 proteins may play a crucial role in DNA demethylation. The essential mechanistic aspects underlying this diverse set of actions are not well understood, primarily due to the lack of reliable structural information on almost all AID/APOBEC family members. None of the quintessential interactions that define specificity to binding partners or susceptibility to antagonistic proteins are well delineated. APOBEC2 and the C-terminal domain of human APOBEC3G are the only members of this class for which high-resolution structures are available. Our work is focused on understanding the structural basis of mechanisms underlying antiviral activity of APOBEC3 proteins and subsequent application of structural information to novel antiviral approaches.
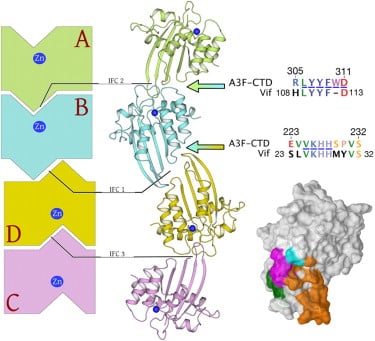
Human APOBEC3F is an antiretroviral single-strand DNA cytosine deaminase, susceptible to degradation by the HIV-1 protein Vif. Recently, we determined the crystal structure of the HIV Vif binding, catalytically active, C-terminal domain of APOBEC3F (A3F-CTD). The A3F-CTD shares structural motifs with portions of APOBEC3G-CTD, APOBEC3C, and APOBEC2. Residues identified to be critical for Vif-dependent degradation of APOBEC3F all fit within a predominantly negatively charged contiguous region on the surface of A3F-CTD. Specific sequence motifs, previously shown to play a role in Vif susceptibility and virion encapsidation, are conserved across APOBEC3s and between APOBEC3s and HIV-1 Vif. In this structure these motifs pack against each other at intermolecular interfaces, providing potential insights both into APOBEC3 oligomerization and Vif interactions (Bohn et al., 2013).
|
|
|
Crystal structure of APOBEC3F domain reveals binding surface of HIV-1 vif. Sequences of crystallographic interfaces conserved across ABOBEC3 family. Crystallographic interfaces maybe functional oligomerization interfaces of ABOPEC3F. HIV-1 Vif has homologous sequences suggesting potential evolution of biomimicry (Bohn et al., 2013). |