HIV Protease Inhibitors
Synthesis and SAR Studies of HIV-1 Protease Inhibitors
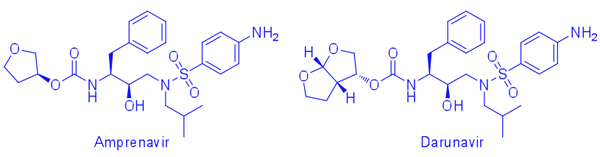
We employ the substrate-envelope model to rationally design protease inhibitors against clinically relevant multidrug-resistant HIV-1 variants. Substrate-envelope model is a general structure-based strategy developed by the Schiffer laboratory that incorporates substrate envelope constraints into structure-based design. In collaboration with Bruce Tidor (MIT) and Tariq Rana (Sanford-Burnham) groups, we have successfully applied the substrate-envelope model to design several libraries of new HIV-1 protease inhibitors based on the (hydroxyethylamino)sulfonamide scaffold. (Ali, et.al., 2010, Nalam, et.al., 2010, Jorissen, et.al., 2009, Altman, et.al., 2008, Reddy, et.al., 2007, Chellappan, et.al., 2007, Ali, et.al., 2006)
The new inhibitors were synthesized and tested using high throughput enzymatic and antiviral assays. These collaborative efforts have led to the discovery of highly potent protease inhibitors that retain potency against clinically relevant multidrug-resistant protease variants. The Monogram Biosciences’ PhenoSenseTM assays demonstrated that these inhibitors were highly effective against a diverse panel of wild-type and drug-resistant viruses. Further development of these protease inhibitors may lead to more effective treatments against drug-resistant HIV-1.
In a recent study, we designed highly potent HIV-1 protease inhibitors (PIs) using the substrate envelope model, which confines inhibitors within the consensus volume of natural substrates, providing inhibitors less susceptible to resistance because a mutation affecting such inhibitors will simultaneously affect viral substrate processing. The designed PIs share a common chemical scaffold but utilize various moieties that optimally fill the substrate envelope, as confirmed by crystal structures. The designed PIs retain robust binding to MDR protease variants and display exceptional antiviral potencies against different clades of HIV as well as a panel of 12 drug-resistant viral strains. The substrate envelope model proves to be a powerful strategy to develop potent and robust inhibitors that avoid drug resistance (Nalam et al., 2013).
 |
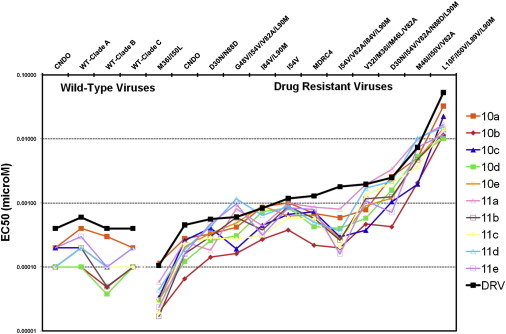 |
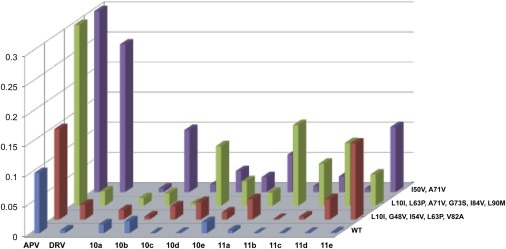
| Binding Affinities of APV, DRV, and the Ten Designed Protease Inhibitors to WT and Drug-Resistant Variants of HIV-1 Protease, determined by a FRET-based enzymatic assay; Ki values are the average of at least three independent measurements. | Resistance Profiles of DRV and the Ten PIs that were DesignedAntiviral potencies (EC50 values) were obtained for WT HIV from clades A, B, and C, and 12 different patient-derived drug-resistant variants of the virus. |

