
Melissa J. Moore, PhD, Howard Hughes Medical Institute Investigator and Professor of Biochemistry and Molecular Pharmacology at UMMS.
Making a movie at the molecular level? A new method of imaging molecule-sized machines as they do the complex work of cutting and pasting genetic information inside the nucleus is the subject of a just-published paper in the journal Science, and the movies have revealed a surprise about how the process works: these critical assembly pathways are a two-way street. This new knowledge could help scientists better understand how the process works in normal cells—and what can go wrong in cancer cells.
Melissa J. Moore, PhD, Howard Hughes Medical Institute Investigator and professor of biochemistry & molecular pharmacology, with colleagues at Brandeis University and Columbia University, has long been studying the spliceosome, a complex of specialized RNA and protein subunits that acts as molecular scissors and tape during gene transcription. The spliceosome snips out the nonsensical sections of genes and stitches the remainder back together to create the final cellular blueprints for making every protein.
Without this cutting and pasting—called RNA splicing—the amount of genetic information that each gene could contain would be strikingly limited. Because of alternate splicing patterns that produce alternate blueprints, each one of our genes can code for multiple proteins; hence the critical importance of the spliceosome.
“Understanding how these micro-machines function inside the cell is important for many reasons,” said Aaron A. Hoskins, PhD, a post-doctoral fellow in Dr. Moore’s lab and first author of the Science paper. “One is to further decipher basic biology—what makes us humans—and another is to understand how diseases related to these different machines come about.”
The spliceosome is so complex that the only way to really understand how it works is to “crack open the hood and take a look,” according to Moore. Enter an innovative new imaging approach that uses lasers, fluorescence microscopy and a method of attaching colorful dyes to the proteins that make up the spliceosome, letting scientists watch the assembly of the spliceosome in motion, in real time. It’s the difference between looking at a photograph of a busy intersection and a movie of the same intersection: objects that might seem rooted in place in the photo—pedestrians waiting to cross—flow in different directions in a film and can be followed along their different paths by the viewer.
The paper, “Ordered and Dynamic Assembly of Single Spliceosomes,” appears in the March 11 issue of Science and caps a five-year collaboration of three research laboratories with remarkably diverse expertise. Members of the laboratory of Virginia Cornish, PhD, in the Department of Chemistry at Columbia University synthesized the special dyes that were attached to the spliceosomal proteins. Jeff Gelles, PhD, the Aron and Imre Tauber Professor of Biochemistry and Molecular Pharmacology at Brandeis University, is an expert in the enzymes necessary to drive tiny molecular motors like the spliceosome; his laboratory at Brandeis developed the multilaser imaging system used to make the movies—and discoveries—in this paper.
The advance of being able to watch spliceosome assembly in real time gives scientists a striking new tool to help understand molecular processes; what Moore and colleagues found in using it is especially surprising. Studying spliceosomes in yeast cells, they found that in the assembly pathway, “what was generally thought to be a one-way street, where assembly takes place step by step in one direction, is actually a two-way street, with lots of places that are dead ends,” said Moore.
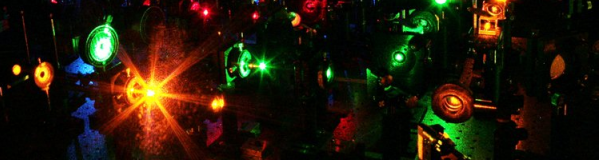 The multilaser prototype used to image spliceosome assembly in real time, housed at Brandeis University. photo credit: Diana Katharine Hunt |
That spliceosome assembly happened in an orderly fashion was understood, but that every step in the process turns out to be reversible was not. (In the busy intersection analogy, two photos taken 15 minutes apart might show the same pedestrian waiting to cross the street, but only a movie would show that she returned to her office to retrieve her umbrella and then came back to the corner in between photos.) As importantly, the movies also showed that early spliceosome assembly steps don’t necessarily mean that the process has started and will move irrevocably forward. Rather “commitment”—the likelihood that the assembly process will be completed once it starts—increases as the process proceeds.
The existence of reversible steps in spliceosome assembly, the dead ends that can stop assembly before it is complete and the increase in commitment as assembly progresses, are now steps that can be seen in real time in the laboratory. Thus one of the most crucial and least understood genetic processes—that of alternative splicing—is suddenly more understandable.
A single human gene has to contain the blueprints for more than one protein. If this weren’t true, the human genome would have to be many times larger than it is. That genes code for multiple proteins explains how all of the genetic information necessary to grow an organism is small enough to be contained inside each one of its cells. This is alternative splicing, in which the RNA produced during gene transcription is reconnected in multiple ways and sets up coding for multiple proteins.
How many? In humans, greater than 95 percent of genes are alternatively spliced by the spliceosome. “Now we have the power to film this molecular tailor in action as it snips and sews scraps of genes to create different protein outfits as required by diverse cell types to carry out their specific functions,” said Moore.
“Dr. Moore’s characterization of the dynamics of assembly of the splicing complex is a remarkable accomplishment and opens new avenues for studying this critical process that differs between normal and cancer cells,” said Phillip A. Sharp, PhD, 1993 Nobel Laureate and Institute Professor at the David H. Koch Institute for Integrative Cancer Research at the Massachusetts Institute of Technology.
“This work has numerous implications for biology and medicine. Splicing is such a fundamental step in the dogmatic transfer of information from DNA to RNA to protein that a deeper understanding is bound to lead to better understanding of when and how this part of the process might go wrong,” said Terence R. Flotte, MD, theCelia and Isaac Haidak Professor in Medicine, executive deputy chancellor, provost, and dean of the School of Medicine. “It is always important when one of the fundamental processes in nature is clearly understood for the first time. This paper is likely to be looked upon as one such landmark.”
Lasers and colors
From neurosurgery to bar code readers, lasers have been used in myriad applications since they were first introduced in the late 1950s. Colleagues in Jeff Gelles’ lab at Brandeis painstakingly developed a way to use lasers to study the spliceosome and its actions.
“The thing that’s very exciting about this technology is that it’s generally applicable to study a wide range of biological problems,” said Dr. Gelles. “It really enables us to find things out that were very difficult to study using previously existing approaches.”
Larry Friedman, PhD, a senior scientist in the biochemistry department at Brandeis, spent more than five years developing specialized light microscopes to watch single protein molecules, while Dr. Hoskins and other co-authors on the Science paper have been developing the methodology to study these proteins in the complex environments necessary for spliceosome function. Dr. Friedman says that there are easily a hundred or so components that comprise the microscope that he designed and built with his colleagues. There are so many parts, in fact, that the microscope is housed on an area that looks much like a billiard table, with small tower-like structures and glass lenses scattered throughout.
To view the spliceosome in action—how it assembles to actually do the splicing—the single yeast components are tagged with florescent dyes then the sample is placed into the microscope. The lasers act as a light source that causes individual tagged molecules to light up so one can actually watch, in unprecedented detail, the splicing process through its various stages.
“If we have one component of the spliceosome that has a green dye on it and one that has a red dye on it, then we see a green spot and a red spot coming together on the RNA, we know that we are studying part of that assembly process,” said Gelles. “By looking at individual molecules one at a time we can actually follow the stages of the assembly process. We can determine whether it happens in the same order on each molecule, or if some spliceosomes assemble differently than others.”
Related links
Science - Ordered and Dynamic Assembly of Single Spliceosomes